Interpretable surface-based detection of FCDs - a MELD study
Machine learning has the potential to revolutionize the field of diagnostic biomedical imaging, but one outstanding challenge is algorithm interpretability. This is particularly important when it comes to incorporating AI for FCD detection into clinical practice.
FCDs are difficult to visualize but often amenable to surgical resection. We wanted to create a robust machine-learning algorithm that could detect FCDs on heterogeneous structural MRI data from epilepsy surgery centres worldwide. However, crucially we wanted to “open the black box” and ensure that the algorithm was interpretable. Clinicians need to be able to understand why the AI identified a particular area.
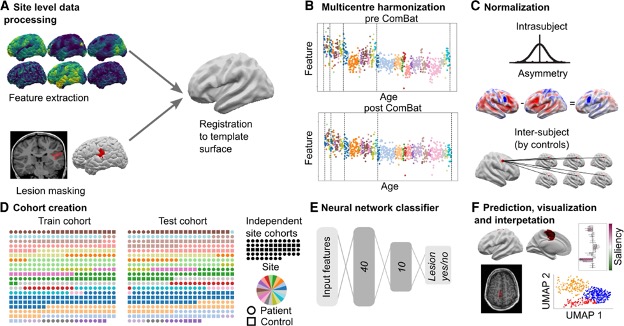
In work published in BRAIN in 2022, the Multi-centre Epilepsy Lesion Detection (MELD) Project set out to develop an open-source, interpretable, surface-based machine-learning algorithm to automatically identify FCDs.
We collated a retrospective MRI cohort of 1015 participants, including 618 patients with focal FCD-related epilepsy and 397 controls, from 22 epilepsy centres around the world. Using 33 surface-based features, we trained and cross-validated a neural network on 50% of the total cohort. We then tested the network on the remaining withheld 50% of the cohort, as well as on two independent test sites.
We used multidimensional feature analysis and integrated gradient saliencies to interrogate network performance.
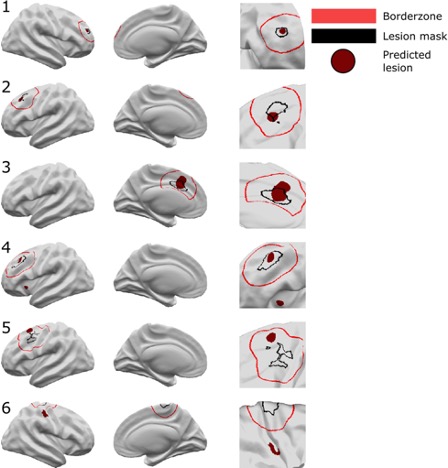
Results
Across the entire withheld test cohort,after including a border zone around lesions to account for uncertainty around the borders of manually delineated lesion masks, the sensitivity was 67%. On a restricted “gold-standard” subcohort of seizure-free patients with FCD type IIB who had T1 and fluid-attenuated inversion recovery MRI data, the MELD FCD surface-based algorithm had a sensitivity of 85%. Specificity was 54%.
Here are examples of classifier performance:
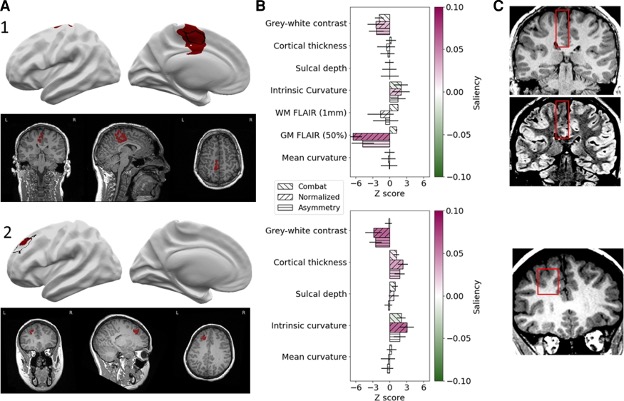
Running the pipeline on new patients
Our pipeline outputs individual patient reports with the location of predicted lesions, along with their imaging features and relative saliency to the classifier. The pipeline also maps the predicted lesions back to the native T1 so that they can be reviewed by a radiologist.
The MELD classifier can be run on MRI data from any 1.5T or 3T scanner on any patient who is over age 3!
In April 2023 (8 months after publication), the MELD team had run 2 workshops to train clinicians and researchers how to use the classifier. We have issued 53 site codes from 35 epilepsy centres.
Overall, by leveraging the power of machine learning and combining it with “explainable AI”, we provide an open-source algorithm for the detection of focal cortical dysplasias.
There are many more cool analyses and results in the paper - so do check it out!
Written with the assistance of ChatGPT